The Evidence & Conclusion Ontology (ECO) is a controlled vocabulary that describes scientific evidence, which results from a variety of research methods, as well as interpretations by authors and scientific curators. ECO is used to document scientific evidence to support conclusions that result from a scientific investigation. Since 2010, ECO has been under the stewardship of scientists at the Institute for Genome Sciences (IGS), University of Maryland, School of Medicine, in Baltimore, Maryland, USA. For a detailed description of ECO architecture and development history, see here.
-
Background
Background
-
History
History
-
License
License
-
Contact Us
Contact Us
-
Staff
Staff
-
Our Users
Our Users
Background
Knowledge Repositories
Like most scientific disciplines, the field of biology is ever expanding and advancing. New research methods are continually developed and a never-ending stream of scientific publications continues to grow. As our scientific knowledge and data associated with such knowledge have increased, mechanisms for representing and storing such information have become essential.
With the growth of the internet & commensurate gains in storage capacity, biological databases have become important repositories of our knowledge. They allow the worldwide community of researchers to access, search, sort, and share a vast amount of information in a way previously not possible. Biological databases can store data but also knowledge learned through scientific analysis of that data. For example, a database might store primary information such as a protein’s amino acid sequence – but it might also contain information that describes that protein’s function, for example “protein X performs some biological function Y”. The amount of information stored by biological databases is immense, interconnected, and complex.
Onotologies for People & Computers
To facilitate efficient storage, search, and retrieval of such information, it is important to represent information in a coherent, consistent, and logical fashion. Such information must be interpretable by computers, but it must also be intuitive and coherent to people who want to access the information. Ontologies allow both people and computers to understand and organize information in such a logical and structured fashion. Thus, scientific concepts at biological databases are represented by ontologies and other controlled vocabularies.
The Evidence & Conclusion Ontology
The Evidence & Conclusion Ontology (ECO) is one such ontology. ECO is used to represent scientific evidence in biological research. Documenting the evidence that supports a scientific assertion such as a protein annotation is essential and fundamental to the scientific method: it allows us to say why we believe what we believe to be true. It also affords us a practical means by which to employ quality control measures when importing data into databases. We can also draw inferences about our confidence in conclusion by looking at associated evidence.
ECO describes evidence arising from laboratory experiments, computational methods, curator inferences, and other means. ECO is currently used by dozens of databases, software tools, and other applications to provide structure and context for documenting evidence in scientific research. Using ECO allows users to query, manage, and interpret data in ways heretofore not possible. Therefore, many of the largest biological and genomic databases in the world already use ECO for summarizing evidence in scientific investigations. But the goal of ECO is to enable summary descriptions of evidence types for a broad range of scientific disciplines. The possibility of expanding ECO to enable representing evidence in disciplines as diverse as anthropology, biodiversity, psychology, and clinical research is being explored.
ECO development is facilitated by National Science Foundation DBI award number 1458400.
History
Origin of ECO
The Evidence Ontology (ECO) was initially created by founders of the Gene Ontology (GO). In tandem with the earliest GO evidence codes, some 100 terms were added to ECO. Today, there are nearly 300 ECO terms, and the approximately 20 GO evidence codes are now mapped to a subset of these ECO terms. The GO uses these evidence terms in the process of annotation, whereby an attribute is assigned to a gene product. GO annotation consists of assigning a molecular function, biological process, or cellular component to a gene product. This information is stored inside a so-called gene association file (GAF), along with a reference, an evidence code, and several other types of information.
For the GO, the purpose of storing evidence is to enable anyone who wants to know why a particular biological attribute was assigned to a particular gene product to know the methods that were employed. That is, a gene product was assigned a particular characteristic by an annotator after a particular methodology was used and an inference was drawn. Incidentally, this is why nearly all the terms in ECO at one point began with the text inferred from followed by the name of a particular research method or result.


-
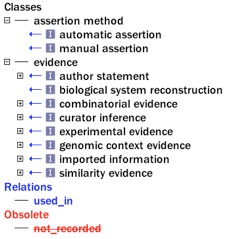
A sample of the set of ECO terms that map to Gene Ontology evidence codes, highlighted in blue.
Early Revisions
In late 2010, an effort was begun to address inconsistencies that existed in ECO, which until recently had been developed in an ad hoc fashion. The following are some updates to the ontology:
- Clarifying what ECO represents & addressing multiple meanings of the word "evidence"
- Defining the main root class evidence
- General structure improvement & moving inappropriately classed terms to appropriate subclasses
- Reviewing term names (labels), correcting misspellings, and generally improving syntax
- Creating new terms requested by users
- Making obsolete any terms that are not evidence, such as not recorded
It was established that evidence is "a type of information that is used to support an assertion" where an assertion is a statement of fact about a thing. This critical piece of information enabled ECO developers to distinguish between evidence and assertion method, both of which were represented as types of evidence in ECO. The latter is actually not a type of evidence, but rather the way in which an assertion (a statement about a thing, an annotation, et cetera) is associated with the thing that the assertion describes. The Gene Ontology term inferred from electronic annotation was renamed automatic assertion. This term was moved to become a subclass of a newly created term assertion method, which is defined as "a means by which a statement is made about an entity". Assertion method distinguishes between human- and computer-based association of statements with the things they describe. Evidence still describes the underlying support for that statement. Both classes can be combined to make complex statements that describe both evidence and assertion method.
Harmonizing with OBI
In 2011, the Evidence Ontology began collaborating with the Ontology for Biomedical Investigations (OBI) in order to better integrate the two ontologies. OBI is particularly well suited to describing instrumentation and research protocols, and it is generally more expressive and detailed than ECO. Many users of ECO desire simple representation of evidence, given their particular database needs, but they often use complex workflows involving multiple methodologies in order to generate the evidence that supports their conclusions. This represents an ontological problem of sorts, because it is often confusing when multiple methods are represented within one ontology term, if they are inconsistently incorporated. Fortunately, complex workflows can be modeled in OBI, and represented as simpler concepts that can be imported into ECO. Work on associating these two ontologies is ongoing.
License
ECO is released into the public domain under CC0 1.0 Universal (CC0 1.0). Anyone is free to copy, modify, or distribute the work, even for commercial purposes, without asking permission. Please see the Public Domain Dedication for an easy-to-read description of CC0 1.0 or the full legal code for more detailed information. To get a sense of why ECO is CC0 as opposed to licensed under CC-BY, please read this thoughtful discussion on the OBO Foundry GitHub site.
Contact Us
We welcome your questions and comments. We use github to track communication for the ECO project. Please click below to submit a github issue with your question, comment, or term suggestion.
Staff

Michelle Giglio, Ph.D.
Principal Investigator

Suvarna Nadendla
Ontologist

Dustin Olley
Website Designer
Project Alumni

Marcus Chibucos, Ph.D.
Ontologist
(previous PI)

Rebecca Jackson (née Tauber)
Ontologist

Elvira Mitraka, Ph.D.
Ontologist

James Munro, Ph.D.
Ontologist

Shoshannah Ball
Intern

Kimuel Villanova
Intern
Our Users
| Group Name | Logo | What They Do |
|---|---|---|
| Alliance of Genome Resources | A consortium of model organism databases and the Gene Ontology Consortium. | |
| Bgee: Gene Expression Evolution | Bgee is a database to retrieve and compare gene expression patterns between animal species. | |
| Bovine Genome Database | A model organism database. | |
| CACAO | The Community Assessment of Community Annotation with Ontologies (CACAO) is a project to do large-scale manual community annotation of gene function using the Gene Ontology as a multi-institution student competition. | |
| CanImmunother | A database of cancer immunotherapies. | |
| CollecTF Database | CollecTF is a database of transcription factor binding sites (TFBS) in the Bacteria domain. It aims at becoming a reference, highly-accessed database by relying on its ability to customize navigation and data extraction, its relevance to the community, the quality and detail of the stored data and the up-to-date nature of the stored information. | |
| DDIEM: Drug Database for Inborn Errors of Metabolism | A database of therapeutic strategies for diseases. | |
| DisProt | DisProt is a database that collects information about intrinsically disordered proteins and proteins with intrinsically disordered regions. Such proteins do not have a fixed three-dimensional structure but rather are dynamic with structures changing in the cell based on conditions. | |
| Disease Ontology (DO) | The Disease Ontology has been developed as a standardized ontology for human disease with the purpose of providing the biomedical community with consistent, reusable and sustainable descriptions of human disease terms, phenotype characteristics and related medical vocabulary disease concepts through collaborative efforts of researchers. | |
| EMBL-EBI - Complex Portal | The Complex Portal is a manually curated, encyclopaedic resource of macromolecular complexes from a number of key model organisms. | |
| EMBL-EBI UniProt-GOA | The UniProt GO annotation program aims to provide high-quality Gene Ontology (GO) annotations to proteins in the UniProt Knowledgebase (UniProtKB). The assignment of GO terms to UniProt records is an integral part of UniProt biocuration. | |
| European Bioinformatics Institute (EMBL-EBI) | EMBL-EBI provides freely available data from life science experiments, performs basic research in computational biology and offers an extensive user training programme, supporting researchers in academia and industry. | |
| Foundation Twenty-nine | Foundation 29 works to provide tools and resources to identify the causes of rare diseases. | |
| Gene Ontology Consortium (GO) | The Gene Ontology (GO) project is a major bioinformatics initiative to develop a computational representation of our evolving knowledge of how genes encode biological functions at the molecular, cellular and tissue system levels. Biological systems are so complex that we need to rely on computers to represent this knowledge. | |
| HDncRNA: Heart Disease-related Non-coding RNAs Database | A database of non-coding RNAs associated with heart disease. | |
| MHC Restriction Ontology | MHC Restriction Ontology | |
| Ontology for Biomedical Investigations (OBI) | The Ontology for Biomedical Investigations (OBI) project is developing an integrated ontology for the description of life-science and clinical investigations. | |
| Ontology for Microbial Phenotypes (OMP) | Web-based community resource designed to display microbial phenotypes and the methods used to study them. | |
| Open Targets | A resource for drug targets and toxicity predictions. | |
| Open Targets Platform | A platform for therapeutic target identification and validation | |
| PRODORIC | A database of prokaryotic gene regulation. | |
| PSISearch2D | An open-source application that runs query-seeded iterative sequence search for remotely related protein detection. | |
| PathoPhenoDB | A database linking human pathogens to phenotypes. | |
| PhaSePro | A database of proteins driving liquid-liquid phase separation. | |
| Phenoscape | Our overall objective is to create a scalable infrastructure that enables linking descriptive phenotype observations across different fields of biology by the semantic similarity of their free-text descriptions. | |
| PlanGexQ | A software tool for the formalization of planarian gene expression patterns into standard morphologies. | |
| Planarian Anatomy Ontology | The Planarian Anatomy (PLANA) Ontology provides comprehensive description of Schmidtea mediterranea anatomical features, and relationships among them, throughout the life cycle of both the sexually and asexually reproducing biotypes. | |
| Plant Ontology | A community resource consisting of standardized terms, definitions, and logical relations describing plant structures and development stages, augmented by a large database of annotations from genomic and phenomic studies. | |
| Planteome | A plant annotation and ontology resource. | |
| Protein Ensemble Database PED | An open access database for the deposition of structural ensembles of intrinsically disordered proteins (IDPs) | |
| Protein Information Resource (PIR) | The Protein Information Resource (PIR) is an integrated public bioinformatics resource to support genomic, proteomic and systems biology research and scientific studies | |
| ProteomeVis | A web app for exploration of protein properties from structure to sequence evolution across organisms’ proteomes | |
| Pseudomonas Genome DB | The Pseudomonas Genome Database is a resource for peer-reviewed high-quality Pseudomonas aeruginosa PAO1 genome annotation, also facilitating whole-genome comparative analyses with other Pseudomonas strains. | |
| RNASIV | RNA structure in viruses | |
| RefSeqFEs | NCBI's RefSeq Functional Elements resource. | |
| Saccharomyces Genome Database (SGD) | The Saccharomyces Genome Database (SGD) provides comprehensive integrated biological information for the budding yeast Saccharomyces cerevisiae along with search and analysis tools to explore these data, enabling the discovery of functional relationships between sequence and gene products in fungi and higher organisms. | |
| SwissLipids | A lipidomic database. | |
| SynGO | The database of synapse biology. | |
| TB TAB ontology | An ontology for factors affecting tuberculosis treatment adherence behavior in sub-Saharan Africa | |
| TIDB- trained immunity database | A comprehensive database of trained immunity | |
| The Arabidopsis Information Resource (TAIR) | The Arabidopsis Information Resource (TAIR) maintains a database of genetic and molecular biology data for the model higher plant Arabidopsis thaliana . | |
| The Ion Channels Variants Portal | A corpus of data of voltage-gated sodium channel mutations. | |
| UniProt | The mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information. | |
| mimicdb | A repository of information about human dimeric integrins | |
| neXtProt | A human protein knowledgebase. |